In the last couple of posts we’ve looked at how to migrate perfmon data from CSV files into SQL Server (
Migrating perfmon CSV files into SQL Server for analysis) and how to use R to graph Perfmon data (
Simple plot of perfmon data in R).
So, the next obvious step is to combine the two, and use R to graph Perfmon data you’ve imported into SQL Server. It turns out that R makes this pretty simple via the RODBC Package.
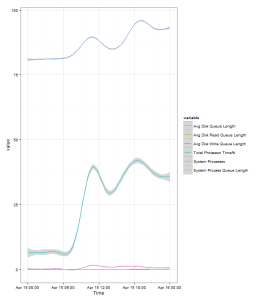
Assuming you’ve created a SQL Server system DSN called perfmon_odbc, then the following code snippet would plot you a lovely graph of the counters:
- % Processor Time
- Avg. Disk Queue Length
- Current Connections
- Processor Queue Length
as recorded on Server A on the 19th June 2013:
install.packages(c("ggplot2","reshape2","RODBC"))
library("ggplot2")
library("reshape2")
library("RODBC")
perfconn<-odbcConnect("perfmon_odbc")
perf<-sqlQuery(perfconn,"select distinct a.CounterDateTime, b.CounterName, a.CounterValue from CounterData a inner join CounterDetails b on a.CounterID=b.CounterID where b.MachineName='\\\\Server A' and b.CounterName in('% Processor Time','Avg. Disk Queue Length','Current Connections','Processor Queue Length') and a.CounterDateTime>'2013-06-19' and a.CounterDateTime<'2013-06-20'")
perf$CounterDateTime <- as.POSIXct(perf$CounterDateTime)
ggplot(data=perf, aes(x=CounterDateTime,y=CounterValue, colour=CounterName)) +
geom_point(size=.5) +
stat_smooth() +
ggtitle("Server A OS Metrics - 11/06/2013")
odbcCloseAll()
Note that this time we didn’t have to melt our data like we did before, as the resultset from RODBC is already in the format we need. But also spot that we’ve had to add some extra \’s to the machine name. \ is an escape character in R, so we need the extra \ to escape the escape character.
Now you’ve all the flexibility of creating the dataset via T-SQL. This means you can start doing things like comparing 2 Servers across the same time period:
install.packages(c("ggplot2","reshape2","RODBC"))
library("ggplot2")
library("reshape2")
library("RODBC")
perfconn<-odbcConnect("perfmon_odbc")
perf<-sqlQuery(perfconn,"select distinct a.CounterDateTime, b.MachineName+' '+b.CounterName as 'CounterName', a.CounterValue from CounterData a inner join CounterDetails b on a.CounterID=b.CounterID where b.MachineName in('\\\\Server A', '\\\\Server B') and b.CounterName in('% Processor Time','Avg. Disk Queue Length','Current Connections','Processor Queue Length') and a.CounterDateTime>'2013-06-19' and a.CounterDateTime<'2013-06-20'")
perf$CounterDateTime <- as.POSIXct(perf$CounterDateTime)
ggplot(data=perf, aes(x=CounterDateTime,y=CounterValue, colour=CounterName)) +
stat_smooth() +
ggtitle("Server A & B OS Metrics - 11/06/2013")
odbcCloseAll()
Note that in this SQL query we’ve specified 2 machine names, and then to make sure that R can distinguish between the counters we’ve appended the Machine Name to the Counter Name in the column list. I’ve also taken out the geom_point(size=.5) line, as with the number of counters now being plotted having the data points make the curves hard to see and compare.
You can extend this to pull the counters for a Server from 2 different days. This makes it easy to check if the spike is a normal daily occurence, or really is the source of your current issues:
install.packages(c("ggplot2","reshape2","RODBC"))
library("ggplot2")
library("reshape2")
library("RODBC")
perfconn<-odbcConnect("perfmon_odbc")
perf<-sqlQuery(perfconn,"select distinct a.CounterDateTime, 'Day 1 -'+b.CounterName as 'CounterName', a.CounterValue from CounterData a inner join CounterDetails b on a.CounterID=b.CounterID where b.MachineName='\\\\Server A' and b.CounterName in('% Processor Time','Avg. Disk Queue Length','Current Connections','Processor Queue Length') and a.CounterDateTime>'2013-06-19' and a.CounterDateTime<'2013-06-20' union select distinct a.CounterDateTime, 'Day 2 -'+b.CounterName, a.CounterValue from CounterData a inner join CounterDetails b on a.CounterID=b.CounterID where b.MachineName='\\\\Server A' and b.CounterName in('% Processor Time','Avg. Disk Queue Length','Current Connections','Processor Queue Length') and a.CounterDateTime>'2013-06-12' and a.CounterDateTime<'2013-06-13'")
perf$CounterDateTime <- as.POSIXct(paste("1900-01-01", substr(perf$CounterDateTime,12,28)))
ggplot(data=perf, aes(x=CounterDateTime,y=CounterValue, colour=CounterName)) +
stat_smooth() +
ggtitle("Server A Metrics - 13/06/2013 and 19/06/2013")
odbcCloseAll()
In this case we union the 2 results sets, and rename the counters to make sure R can identify which set are which date. We also set the date component on the time stamps to be the same day (I use 01/01/1900 as it’s nice and obvious when looking at a chart later to see that it’s been reset), this is to make sure R plots the time values against each other correctly.